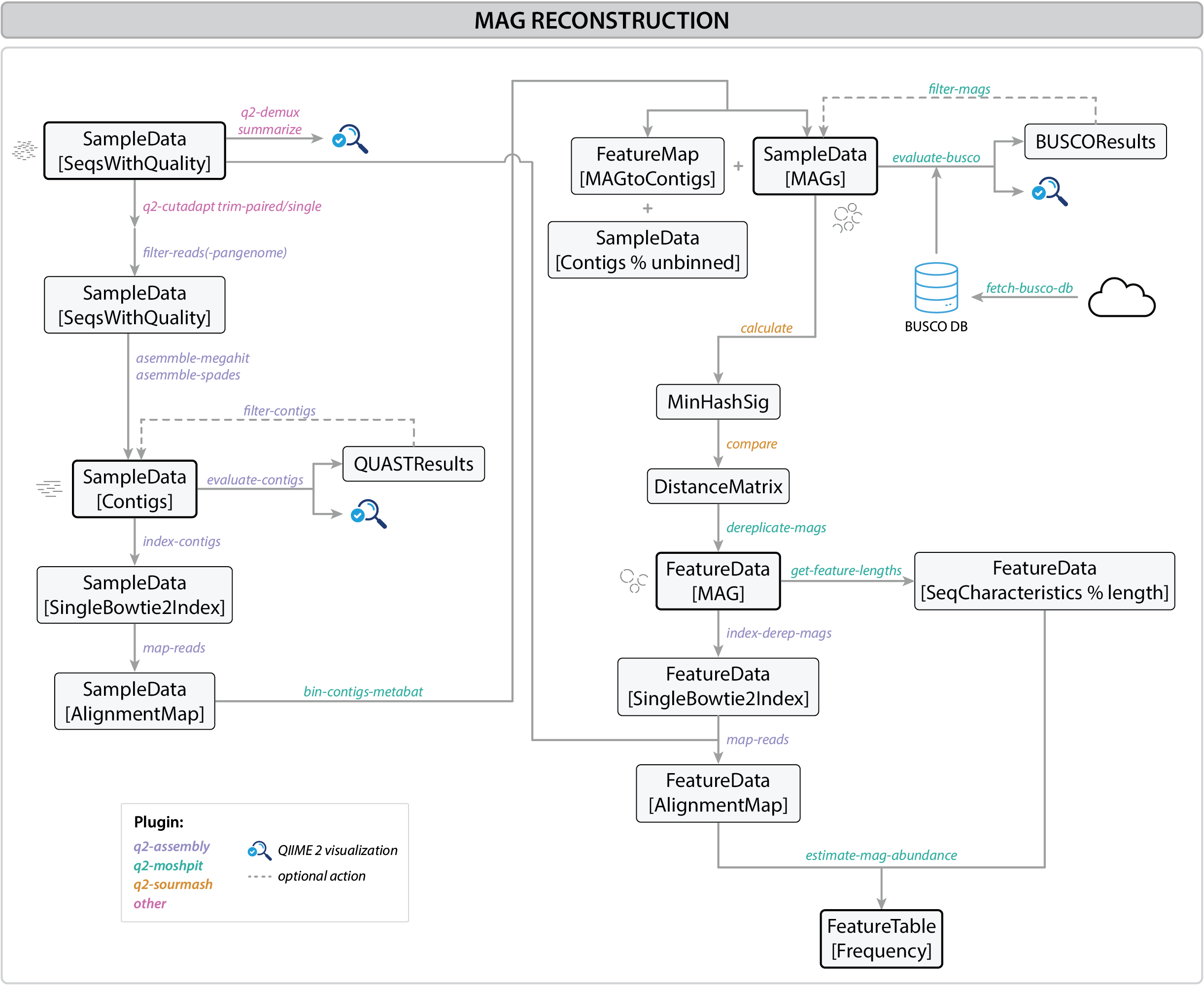
Figure 1:MAG reconstruction workflow
Metagenome-assembled genomes (MAGs) are genomes reconstructed directly from DNA from complex microbial mixtures without the need for culturing organisms in the laboratory. This approach allows researchers to explore the genetic makeup of microbial communities in various environments, providing insights into the diversity, functions, and ecological roles of uncultured microorganisms. Recovering MAGs involves assembling sequencing reads into contigs, binning contigs into draft genomes, and evaluating their quality. Optionally, MAGs can be further dereplicated into a set of non-redundant genomes.
This workflow describes a step-by-step process for MAGs recovery using QIIME2. Each command includes explanations of the parameters used.
For more information on the tools used in this workflow, refer to their official documentation:
- MEGAHIT: https://
github .com /voutcn /megahit - SPAdes: https://
github .com /ablab /spades - QUAST: https://
github .com /ablab /quast - MetaBAT: https://
bitbucket .org /berkeleylab /metabat/ - BUSCO: https://
gitlab .com /ezlab /busco - Sourmash: https://
github .com /dib -lab /sourmash - Kraken2: https://
github .com /DerrickWood /kraken2 - QIIME 2: https://
github .com /qiime2